Axe 1. Régulation de l’expression des gènes et ciblage thérapeutique Responsables : Laurence CHOULIER et Dmytro DZIUBA
Les thématiques associées à l’axe ‘Régulation de l’expression des gènes et ciblage thérapeutique’, visent à la compréhension des mécanismes moléculaires qui sous-tendent la régulation de l’expression des gènes, et le développement de nouveaux vecteurs et stratégies de ciblage basés sur des aptamères.
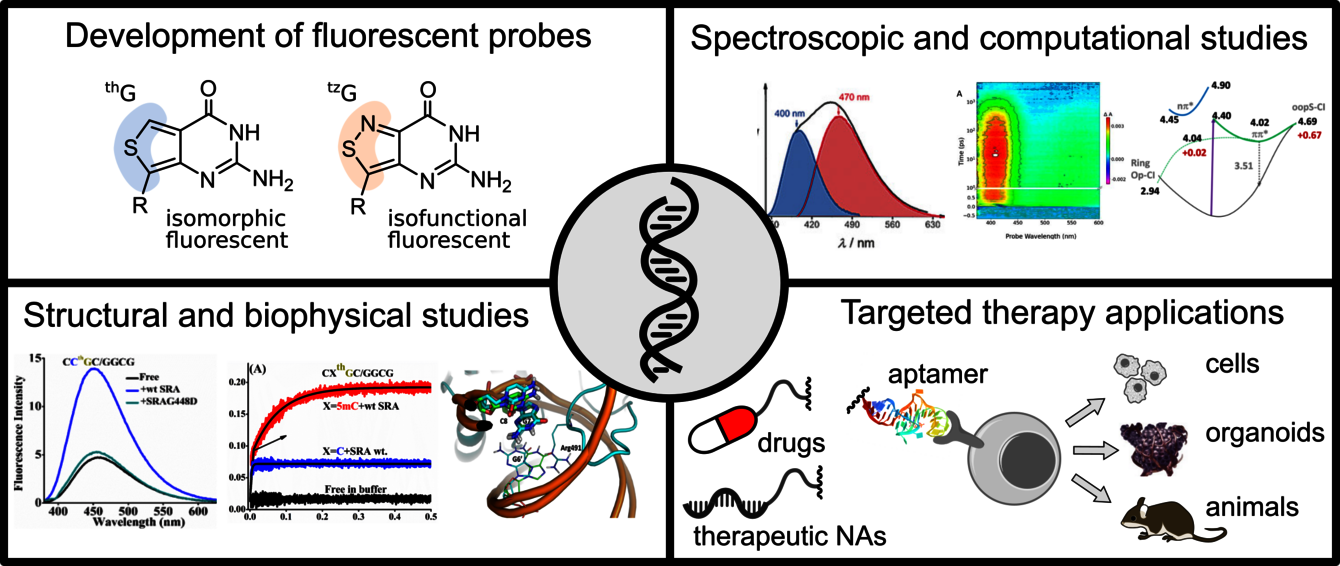
L’une de nos thématiques explore les interactions entre les G-quadruplexes (G4) et les protéines de repliement/dépliement des G4. Les G4, structures non canoniques formées par des tétrades de guanine (G), jouent un rôle clé dans le maintien de l'intégrité des chromosomes et le contrôle de la transcription des gènes. L’originalité de notre approche repose sur la synthèse à grande échelle, la caractérisation photophysique et l’utilisation d'analogues fluorescents de la guanine pour étudier les dynamiques et interactions des G4. La thiénoguanosine (thG) et l’isothiazologuanosine (tzG) sont des mimes fluorescents presque parfaits de guanine, qui permettent de remplacer n’importe quel résidu G dans des séquences d’ARN ou d’ADN. Sensibles à leur environnement, thG/tzG permettent de suivre sélectivement et précisément les changements conformationnels, facilitant ainsi l'observation des interactions protéines/G4. Notre premier objectif est d’étudier finement les mécanismes photophysiques de thG/tzG dans des séquences modèles et des G4 pour en faire des rapporteurs quantitatifs de la structure locale des ADN/ARN. Ces propriétés spectroscopiques seront alors utilisées pour étudier les mécanismes de dépliement des G4 par les protéines DHX36 et CNBP dans des mesures d’ensemble et à l’échelle de la molécule unique. Nous étudions également le rôle des G4 dans la machinerie épigénétique, notamment leur interaction avec les protéines DNMT1 et UHRF1, qui contrôlent la méthylation de l'ADN.
Une autre de nos thématiques vise à promouvoir des stratégies de ciblage, multifonctionnelles, versatiles, à base d’aptamères. Les aptamères sont des ARN ou ADN simple brin. Leur taille (6-30 kDa), leurs propriétés physico-chimiques, leur facilité et faible coût de synthèse, leur caractère non-immunogène, leur affinité et sélectivité pour leurs cibles, en font une alternative pertinente aux anticorps. Ils ont déjà trouvé leur place dans des applications thérapeutiques, mais leur potentiel en tant que molécules d'adressage reste encore sous-exploité. Nous nous intéressons aux aptamères sous forme de conjugués multi-moléculaires ou décorant des nanoparticules. Nous développons des stratégies permettant de comprendre la dynamique du ciblage (affinité/spécificité/avidité/stabilité/transport/libération des principes actifs), et de délivrer spécifiquement différentes molécules (G4, mais aussi des acides nucléiques d’intérêt clinique et drogues) dans des environnements biologiques complexes et/ou au sein des cellules ciblées, et ainsi contribuer à la compréhension demécanismes épigénétiques et au développement des acides nucléiques thérapeutiques. Les applications diagnostiques et pharmaceutiques concernent différents foyers tumoraux, inflammatoires et infectieux.