Axis 1. Regulation of gene expression and therapeutic targeting Leaders: Laurence CHOULIER and Dmytro DZIUBA
The topics covered by the research axis ‘Regulation of gene expression and therapeutic targeting’ aim to understand the molecular mechanisms underlying the regulation of gene expression, and to develop new vectors and targeting strategies based on nucleic acid aptamers.
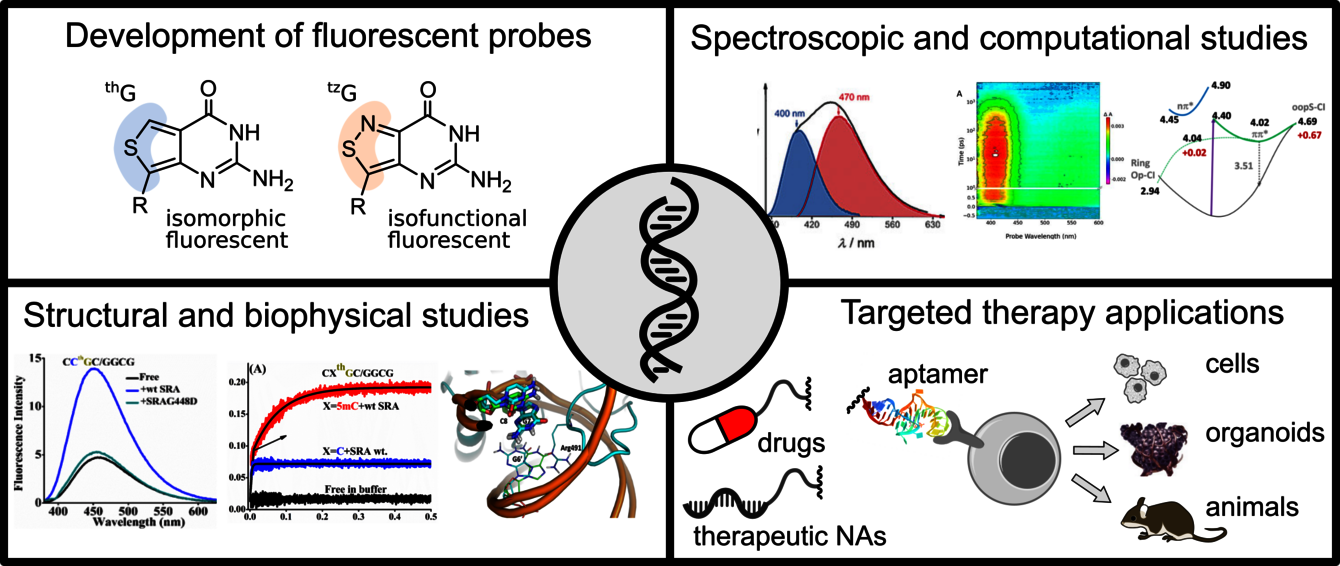
One of our topic explores the interactions between G-quadruplexes (G4s) and G4 folding/unfolding proteins. G4s are non-canonical structures formed by guanine (G) tetrads, which play a major role in the maintenance of chromosome integrity and the control of gene transcription. Our original approach is based on large-scale synthesis, photophysical characterisation and the use of fluorescent guanine analogues to study the dynamics and interactions of G4s. Thienoguanosine (thG) and isothiazologuanosine (tzG) are quasi-perfect fluorescent mimics of guanine, which can replace any G residue in RNA or DNA sequences. Being sensitive to their environment, thG/tzG can be used to selectively and precisely monitor conformational changes, facilitating the observation of protein/G4 interactions. Our first objective is to conduct a detailed study of the photophysical mechanisms of thG/tzG in reference sequences and G4s as quantitative reporters of the local structure of DNA/RNA. These spectroscopic properties will then be used to study the mechanisms of G4 unfolding by the DHX36 and CNBP proteins in overall and single-molecule measurements. We are also studying the role of G4s in the epigenetic machinery, in particular their interaction with the DNMT1 and UHRF1 proteins, which control DNA methylation.
We are also developing multifunctional, versatile targeting strategies based on nucleic acid aptamers. Aptamers are single-stranded RNA or DNA. Their size (6-30 kDa), physicochemical properties, ease and low cost of synthesis, non-immunogenic nature, affinity and selectivity for their targets, make them relevant alternatives to antibodies. They are already used in therapeutic applications, their properties as delivery molecules remain under-exploited. We are interested in using aptamers as multi-molecular conjugates or to decorate nanoparticles. Strategies are being developed to understand the dynamics of targeting (affinity/specificity/avidity/stability/transport/release of active molecules), and to specifically deliver different molecules (G4, but also nucleic acids of clinical interest and drugs) in complex biological environments and/or within targeted cells, thereby contributing to the understanding of epigenetic mechanisms and to the development of therapeutic nucleic acids. Diagnostic and pharmaceutical applications concern various tumour, inflammatory and infectious sites.